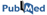Ubiquitination
Ubiquitination is a protein modification in which ubiquitin molecule covalently binds to the lysine residues of ubiquitin substrates (Hicke, 2001). This Two Sample Logo example contains 25 residue fragments, 12 upstream and 12 downstream, from all lysines found in 95 ubiquitinated proteins reported by (Hichcock et al., 2003) and (Peng et al., 2003). The positive sample contains 110 non-redundant fragments around experimentally verified ubiquitination sites, while the negative sample contains all remaining lysines from the same set of proteins, 2885 in total. In order to help answering a question about sequence biases around ubiquitination sites, a two sample logo can be generated to visualize residues that are significantly enriched or depleted in the set of ubiquitinated fragments.
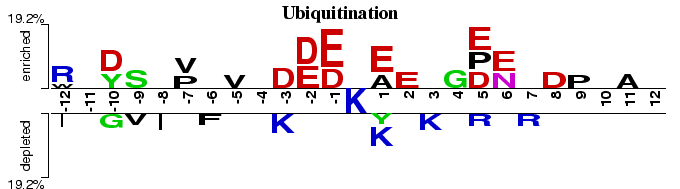
Tyrosine Phosphorylation
Reversible protein phosphorylation provides a major regulatory mechanism in eukaryotic cells. At least one third of eukaryotic proteins is believed to be phosphorylated (Marks, 1996). This Two Sample Logo example contains 25 residue fragments, 12 upstream and 12 downstream, from all tyrosines found in 3408 phosphorylated proteins reported by (Iakoucheva et al., 2004). The positive sample contains 136 non-redundant fragments around experimentally verified phosphorylation sites, while the negative sample contains all remaining tyrosines from the same set of proteins, 5103 in total. In order to help answering a question about sequence biases around phosphorylation sites, a two sample logo can be generated to visualize residues that are significantly enriched or depleted in the set of phosphorylated fragments.
Calmodulin IQ Motifs

Calmodulin signaling involves important and wide-spread eukaryotic protein-protein interactions that regulate a variety of cellular processes (Vetter and Leclerc, 2003). Among five major types of calmodulin binding sites, one particular type of calmodulin regulation is predominantly calcium independent and requires so-called IQ motifs as the calmodulin binding regions. This Two Sample Logo example is based on the Calmodulin Target Database (Yap et al., 2000) and contains a set of 17 non-redundant experimentally verified IQ motifs and 294 remaining fragments, all containing dipeptide IQ, from the same set of proteins. A two sample logo can be generated in order to understand sequence biases that distinguish IQ motifs from the remaining fragments that contain IQ, but are not calmodulin-binding.
Calmodulin Target Database suggests that a regular expression for the calmodulin IQ motif is [FILV]QXXX[RK]GXXX[RK]XX[FILVWY]. However, even though positive and negative sequences in this example were restricted to those starting with an IQ dipeptide only, the two-sample logo reveals interesting dependencies when compared to the negative sites.
Exon-Intron Splice Sites
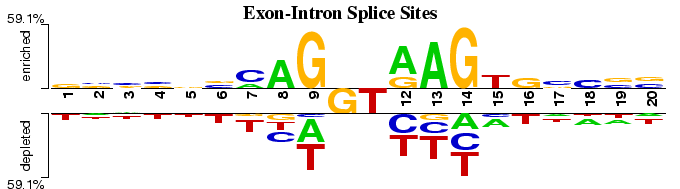
This Two Sample Logo example displays the differences between experimentally verified and false positive exon-intron junction sites, both downloaded from HS3D database (Pollastro and Rampone, 2002). The positive set contains 2,000 non-identical 20-nucleotide long exon-intron junctions centered at GT dinuclotide. The negative set contains 2,000 non-identical randomly selected false positive junctions, also centered at GT. In contrast to a one-sample WebLogo output, this example clearly indicates higher GC content on the intron (3') side of the junction.
Interestingly, both 5' and 3' sides show significant depletion of thymine, except at position 15 (6th base within an intron), where it is significantly enriched in the positive dataset.
Alternatively/regularly spliced exon-intron junctions
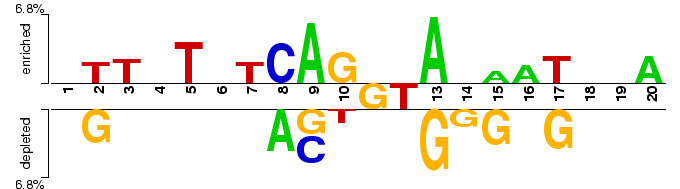
Two sample logo of the differences between alternatively and regularly spliced exon-intron junctions for the p-value threshold of 0.05. A random sample of 2,000 alternatively spliced sites centered at GT dinucleotide (positive sample) was extracted from HASDB (Modrek, et al., 2001) as 20 nucleotide-long sequences around 5’ splice` sites which had more than one competing 3’ site. Regular splice sites (negative sample) consisted of a random sample of 2,000 non-identical exon-intron junctions from HS3D database (Pollastro and Rampone, 2002).
References
- Hicke, L. (2001) "Protein regulation by monoubiquitin." Nat. Rev. Mol. Cell Biol., 2:195-201.
- Hitchcock, A.L., Auld, K., Gygi, S.P., and Silver, P.A. (2003) "A subset of membrane-associated proteins is ubiquitinated in response to mutations in the endoplasmic reticulum degradation machinery." Proc Natl Acad Sci USA, 100:12735-12740.
- Iakoucheva, L.M., Radivojac, P., Brown, C.J., O'Connor, T.R., Sikes, J.G., Obradovic, Z., Dunker, A.K. (2004) "The importance of intrinsic disorder for protein phosphorylation." Nucleic Acids Research, 32(3):1037-1049.
- Marks, F. (1996) Protein phosphorylation. VCH Weinheim: New York.
- Modrek, B., Resch A., Grasso C., and Lee C. (2001) "Genome-wide detection of alternative splicing in expressed sequences of human genes." Nucleic Acids Res, 29, 2850-2859.
- Peng, J., Schwartz, D., Elias, J.E., Thoreen, C.C., Cheng, D., Marsischky, G., Roelofs, J., Finley, D., and Gygi, S.P. (2003) "A proteomics approach to understanding protein ubiquitination." Nat. Biotech., 21:921-926.
- Pollastro, P. and Rampone, S. (2002) "HS3D, a dataset of homo sapiens splice regions and its extraction procedure from a major public database." International Journal of Modern Physics, 13, 1105-1117.
- Vetter S.W., Leclerc E. (2003) "Novel aspects of calmodulin target recognition and activation." Eur J Biochem 270(3):404-414.
- Yap K., Kim J., Truong K., Sherman M., Yuan T., Ikura M. (2000) "Calmodulin target database." J Struct Funct Genomics 1:8-14.